R emmeans package output in Shiny
Q: How to unite interaction columns from R emmeans package dynamically generated in R-Shiny?
I am building an R-Shiny app where I need to wrangle the output from the 'emmeans' package. However, in this interactive environment where many factors may be entered by the user, the single-tibble 'emmeans' output structure will vary with each run depending on the selections made. It could go from having only a single main effect to having multiple 3-way interactions (mixed with main effects and 2-way interactions) arranged in a wide format way.
For instance, assuming the user selects FctrA (with levels A and B) and FctrB (with levels C, D, and E), the interaction FctrA_FctrB will be automatically considered as well. When (~FctrA, ~FctrB, ~FctrA+FctrB) are submitted to 'emmeans', the output tibble is structured as follows:
- the leftmost side of the tibble contains FctrA results (levels, estimates, SE, df, CLs);
- the certermost block contains FctrB results;
- the rightmost side of the tibble contains the interaction results
So far so good except that FctrA levels columns is a single column, FctrB levels column is also a single column, but the interaction portion has its levels split into two columns, one with FctrA and one with FctrB.
The above issue impairs gathering, spreading, stacking of the separate blocks owing to the dimensional discrepancy.
My question is: How can I tell Shiny ('tidyr') to find those split interaction columns and concatenate them appropriately in a smart way (column names, number of interaction columns, overall output tibble structure varies according to input data and selections made)?
I am attaching an image to illustrate the process exemplified above:
- Figure 1 is what I currently have from 'emmeans';
- Figure 2 is what I need to do with any interactions I may have in the tibble (see column 13_14 related to my question above);
- Figure 3 is the final table I need which I believe I may get with the function 'coalesce' as long all blocks in my tibble have the same number of columns
Thank you all in advance for any advice you may provide.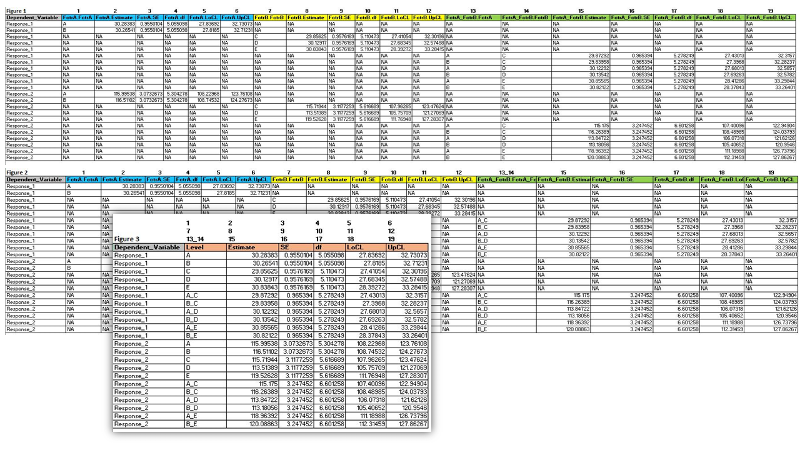
I am building an R-Shiny app where I need to wrangle the output from the 'emmeans' package. However, in this interactive environment where many factors may be entered by the user, the single-tibble 'emmeans' output structure will vary with each run depending on the selections made. It could go from having only a single main effect to having multiple 3-way interactions (mixed with main effects and 2-way interactions) arranged in a wide format way.
For instance, assuming the user selects FctrA (with levels A and B) and FctrB (with levels C, D, and E), the interaction FctrA_FctrB will be automatically considered as well. When (~FctrA, ~FctrB, ~FctrA+FctrB) are submitted to 'emmeans', the output tibble is structured as follows:
- the leftmost side of the tibble contains FctrA results (levels, estimates, SE, df, CLs);
- the certermost block contains FctrB results;
- the rightmost side of the tibble contains the interaction results
So far so good except that FctrA levels columns is a single column, FctrB levels column is also a single column, but the interaction portion has its levels split into two columns, one with FctrA and one with FctrB.
The above issue impairs gathering, spreading, stacking of the separate blocks owing to the dimensional discrepancy.
My question is: How can I tell Shiny ('tidyr') to find those split interaction columns and concatenate them appropriately in a smart way (column names, number of interaction columns, overall output tibble structure varies according to input data and selections made)?
I am attaching an image to illustrate the process exemplified above:
- Figure 1 is what I currently have from 'emmeans';
- Figure 2 is what I need to do with any interactions I may have in the tibble (see column 13_14 related to my question above);
- Figure 3 is the final table I need which I believe I may get with the function 'coalesce' as long all blocks in my tibble have the same number of columns
Thank you all in advance for any advice you may provide.

ASKER CERTIFIED SOLUTION
membership
This solution is only available to members.
To access this solution, you must be a member of Experts Exchange.